
Tianyu Cui
I'm Tianyu, a Senior Research Scientist at Johnson & Johnson in London, working on AI for drug discovery. My research spans Bayesian modeling to modern deep learning, with a particular focus on building principled probabilistic approaches that leverage and adapt foundation models for scientific prediction and discovery. I work across Bayesian modeling, deep generative model, causality, and experimental design to develop models that are not only accurate, but uncertainty-aware, interpretable, and experimentally actionable for real-world decision-making in scientific discovery.
I did my PhD at Probabilistic Machine Learning and Machine Learning for Health group at Aalto University with Prof. Samuel Kaski and Prof. Pekka Marttinen focusing on Bayesian deep learning before joining Imperial College London as a Research Associate. I received MSc degree from University College London on computational statistics and machine learning.
Email | LinkedIn | GitHub | Google Scholar
Selected publications
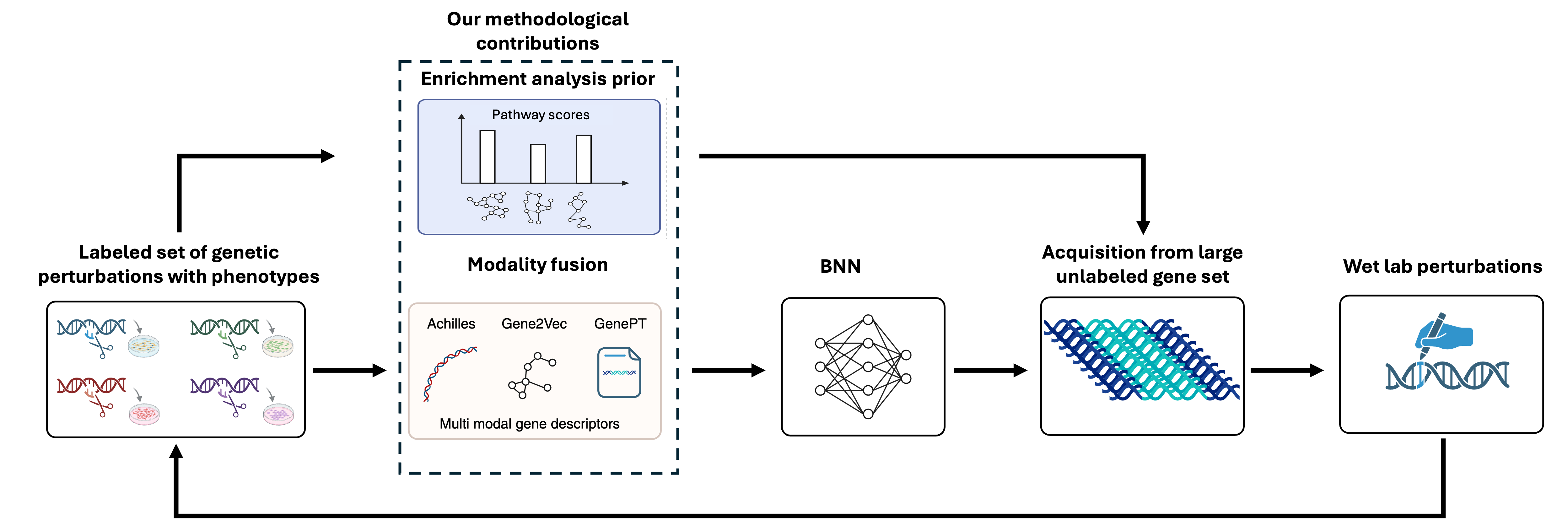
BioBO: Biology-informed Bayesian Optimization for Perturbation Design
SPIGM@ Neural Information Processing Systems (NeurIPS) 2025
We introduce BioBO, a biology-informed Bayesian optimization framework for genomic perturbation design that integrates multimodal gene embeddings with pathway enrichment analysis to guide surrogate modeling and acquisition. By combining biologically grounded priors with principled exploration–exploitation, BioBO improves the ability to identify high-value gene perturbations while providing pathway-level interpretability. Experiments on public CRISPR datasets show that BioBO improves labeling efficiency by 25–40% and consistently outperforms standard BO methods.
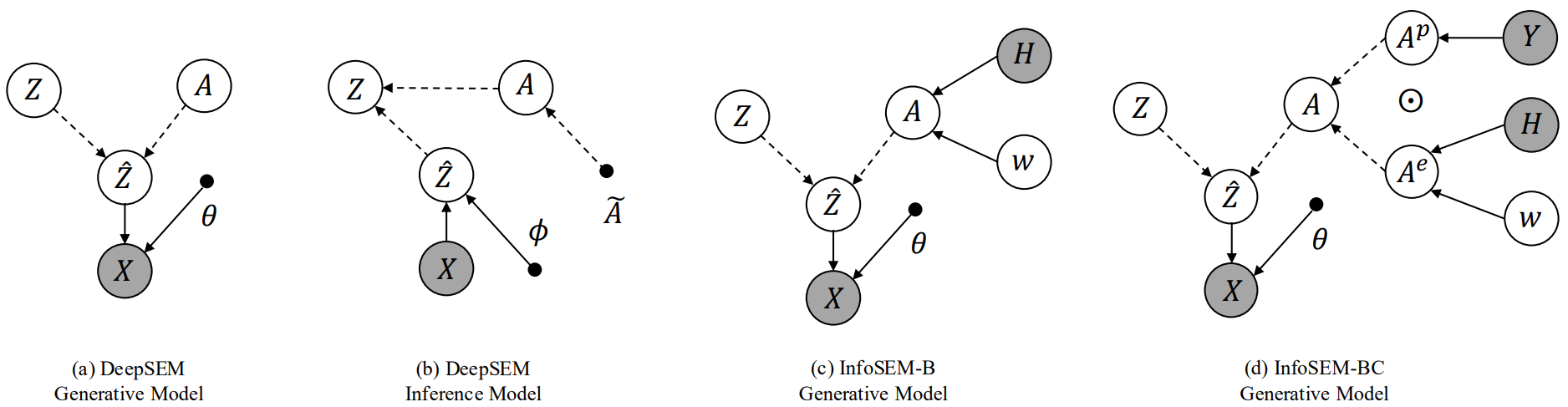
InfoSEM: A Deep Generative Model with Informative Priors for Gene Regulatory Network Inference
International Conference on Machine Learning (ICML) 2025
AI4NA@ International Conference on Learning Representations (ICLR) 2025 (Oral)
We introduce InfoSEM, an unsupervised deep generative model for GRN inference that integrates gene embeddings from pretrained foundation models and known interactions as informative priors. We show that existing supervised methods exploit dataset biases rather than true biological mechanisms. We propose a biologically motivated benchmarking that better reflects real-world applications, where InfoSEM achieves state-of-the-art performance.
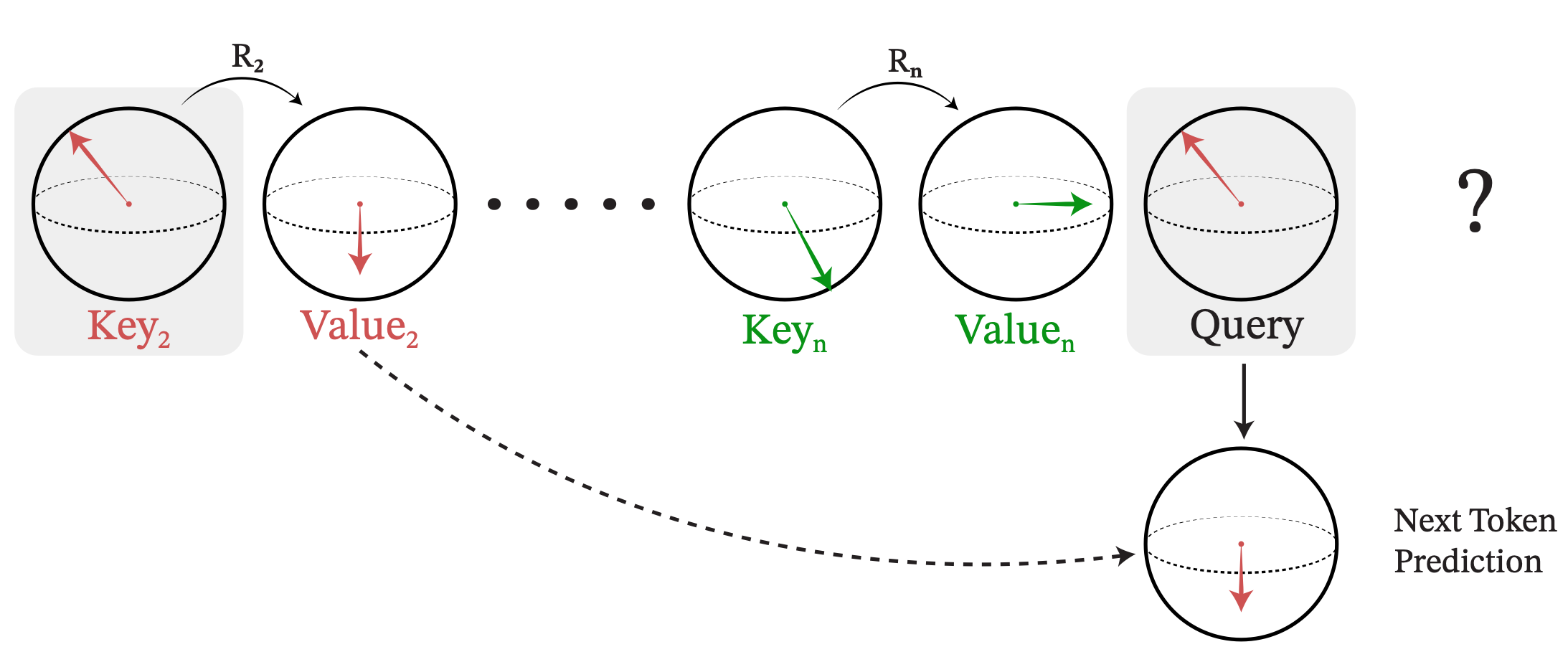
Geometric Hyena Networks for Large-scale Equivariant Learning
International Conference on Machine Learning (ICML) 2025 (Spotlight)
We introduce Geometric-Hyena operator, a translation and rotation equivariant long-convolutional method to process global geometric context at scale with sub-quadratic complexity. Significantly more compute and memory efficient than transformers.
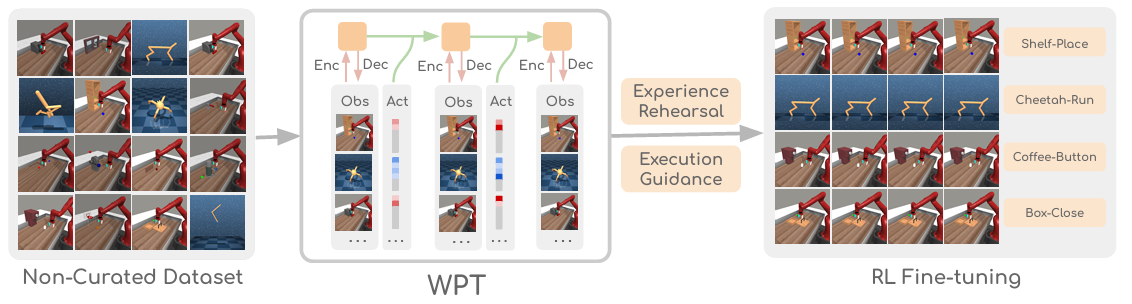
Generalist World Model Pre-Training for Efficient Reinforcement Learning
World Models@ International Conference on Learning Representations (ICLR) 2025
We propose a generalist world model pretraining (WPT) approach for robot learning using reward-free, non-expert multi-embodiment offline data. Combined with retrieval-based experience rehearsal and execution guidance, WPT enables efficient reinforcement learning and rapid task adaptation, outperforming learning-from-scratch baselines by over 35% across 72 visuomotor tasks.
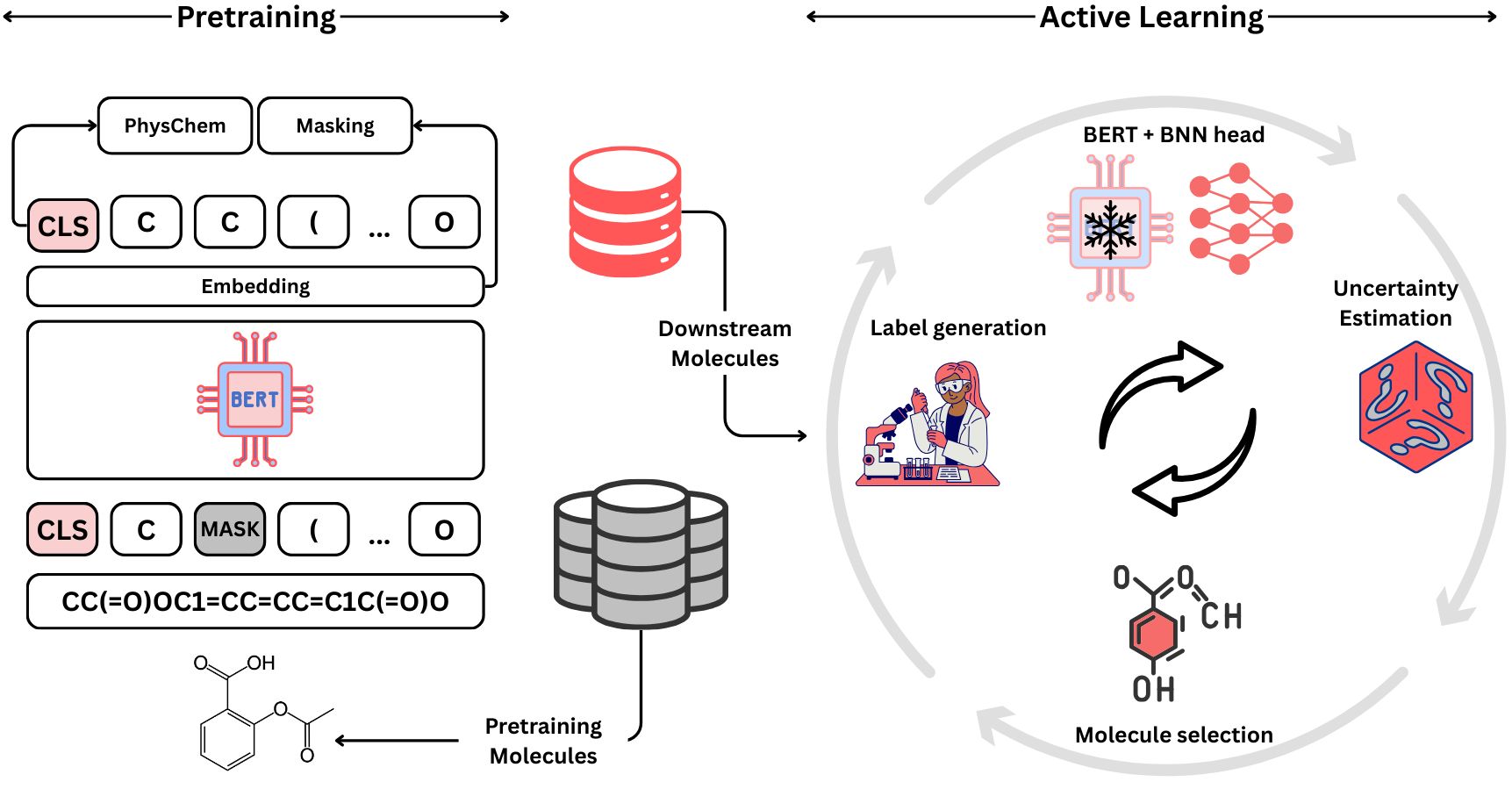
Molecular Property Prediction using Pretrained-BERT and Bayesian Active Learning: A Data-Efficient Approach to Drug Design
Journal of Cheminformatics 2025
GEM@ International Conference on Learning Representations (ICLR) 2025
We propose a semi-supervised Bayesian active learning framework for molecular property prediction, leveraging representations from a pretrained MolBERT model on 1.26 million compounds instead of relying solely on limited labeled data. This semi-supervised approach improves prediction accuracy and acquisition efficiency compared to classical supervised active learning.
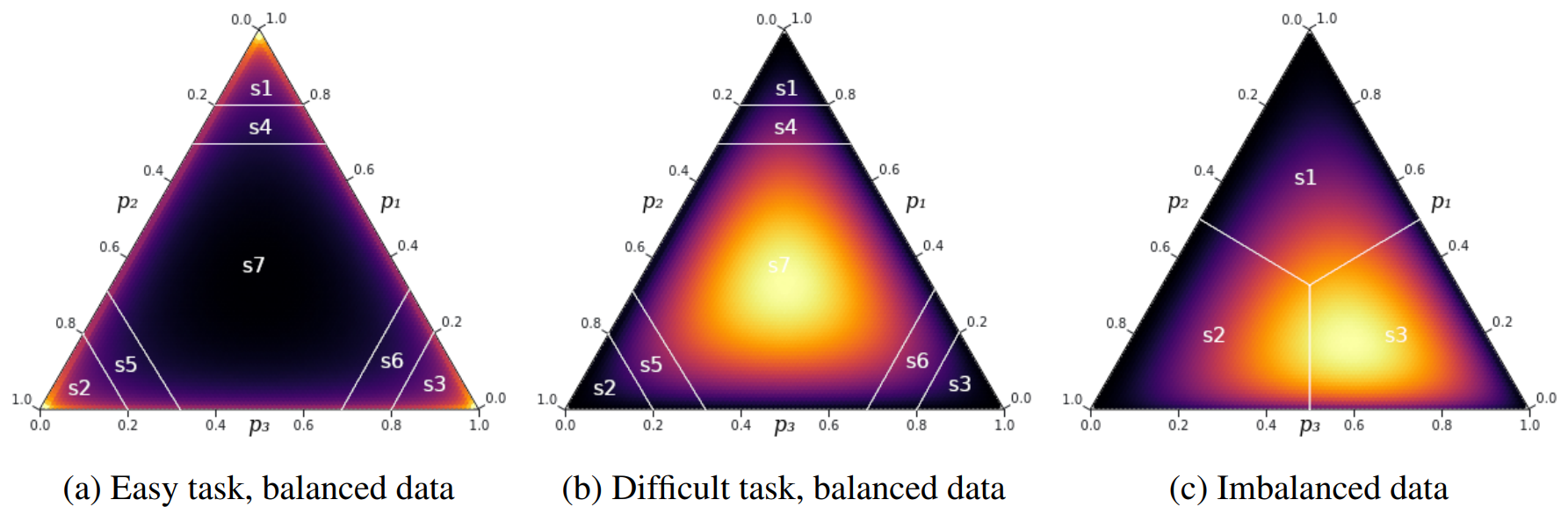
Incorporating Functional Summary Information in Bayesian Neural Networks Using a Dirichlet Process Likelihood
International Conference on Artificial Intelligence and Statistics (AISTATS) 2023
We introduce DP-BNN, a novel strategy that integrates prior knowledge of task difficulty and class imbalance into deep learning with a Dirichlet process on the predicted probabilities. DP-BNN improves in accuracy, uncertainty calibration, and robustness against corruptions on both balanced and imbalanced image and text datasets with negligible computational overhead.
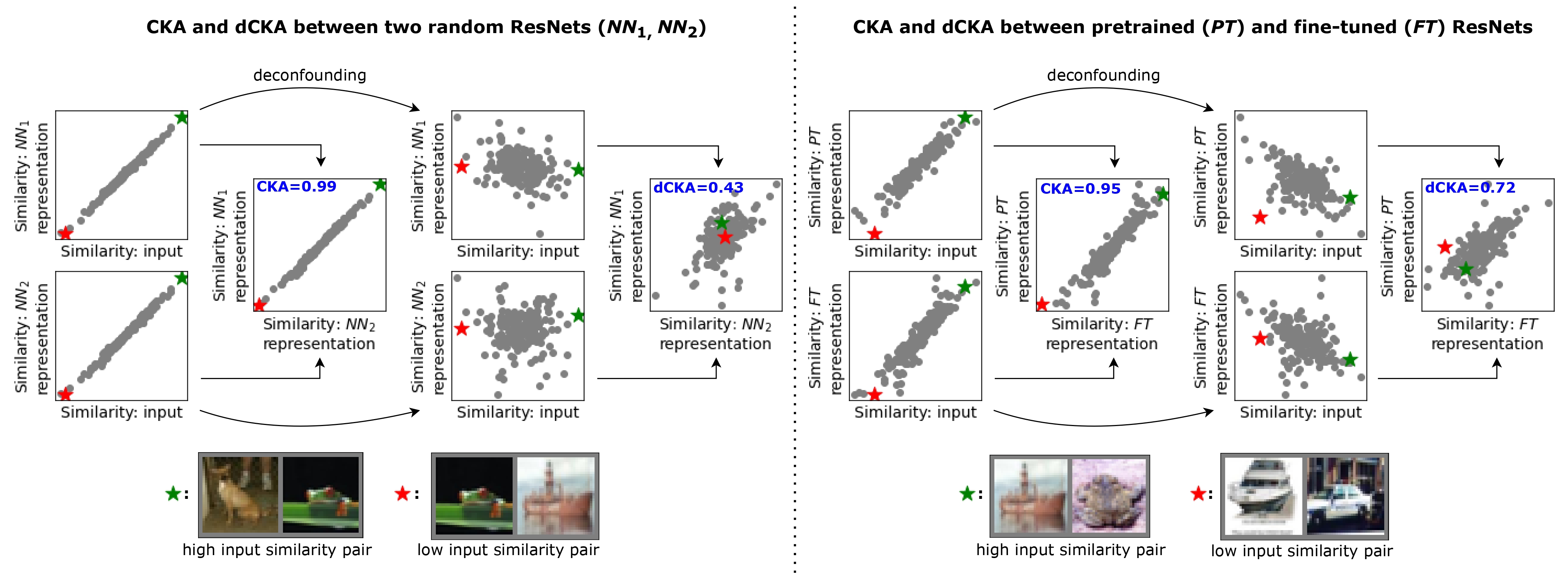
Deconfounded Representation Similarity for Comparison of Neural Networks
Neural Information Processing Systems (NeurIPS) 2022 (Oral)
Representation similarity between neural networks can be confounded by population structure, leading to misleading conclusions, such as spuriously high similarity in random neural networks. We propose covariate adjustment regression to remove this confounder, enhancing functional similarity detection while preserving invariance properties.
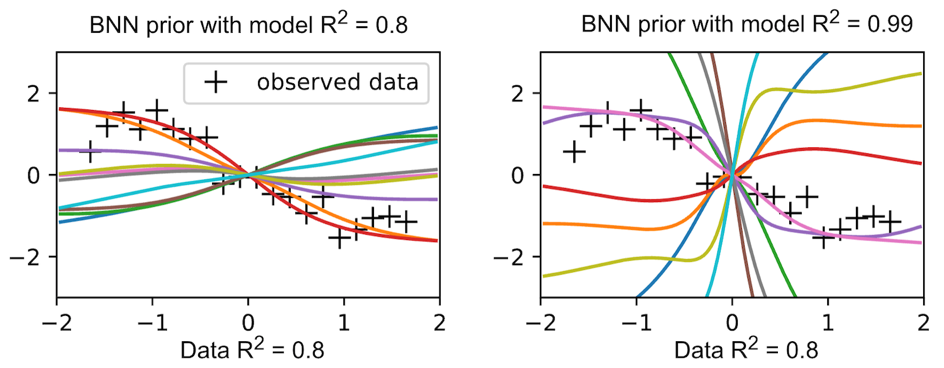
Informative Bayesian Neural Network Priors for Weak Signals
Bayesian Analysis 2022
Joint Statistical Meetings 2022 (Oral)
We introduce a novel framework that incorporates domain knowledge on feature sparsity and data signal-to-noise ratio into the Gaussian scale mixture priors of neural network weights. This informative prior enhances prediction accuracy in datasets with weak and sparse signals, such as those in genetics, even surpassing computationally intensive cross-validation for hyperparameter tuning.
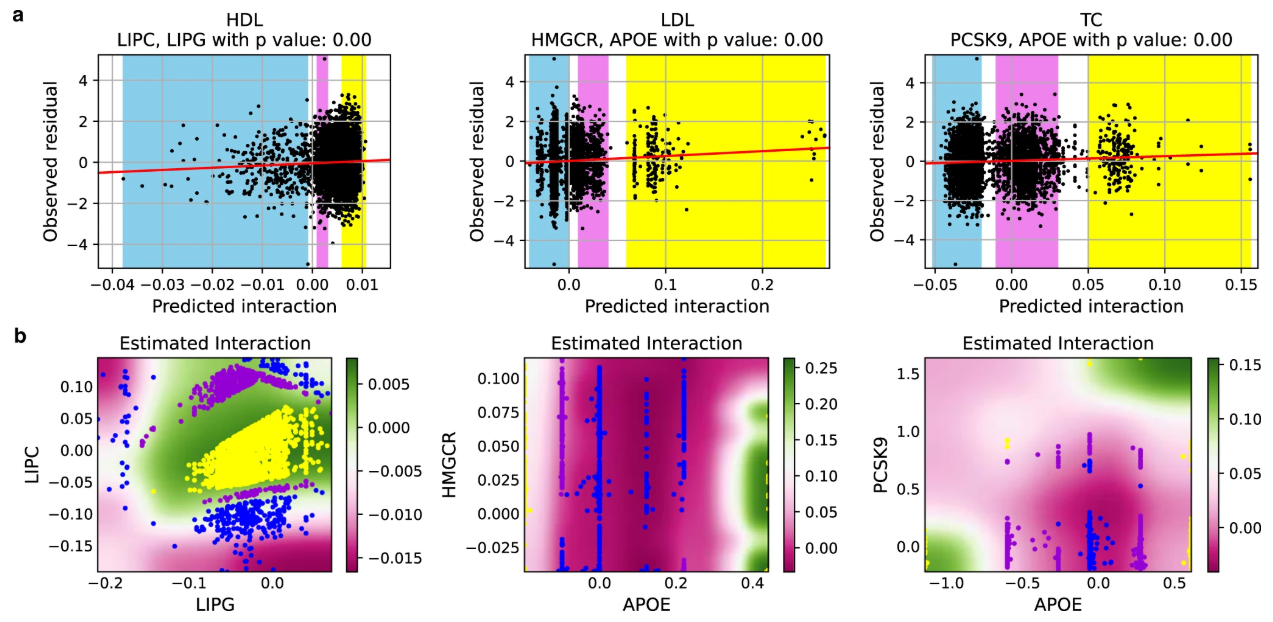
Gene-gene Interaction Detection with Deep Learning
Nature Communications Biology 2022
We propose a biologically motivated neural network architecture to model and detect gene interactions from GWAS datasets, along with a novel permutation procedure to assess the uncertainty of these interactions. The proposed framework identified nine interactions in a cholesterol study using the UK Biobank, which were successfully replicated in an independent FINRISK dataset.
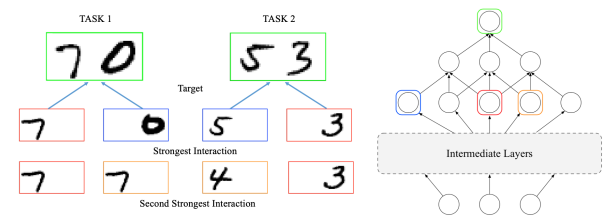
Learning Global Pairwise Interactions with Bayesian Neural Networks
European Conference on Artificial Intelligence (ECAI) 2020 (Oral)
BDL@ Neural Information Processing Systems (NeurIPS) 2019
In this paper, we propose using Bayesian neural networks to capture global interactions effects with well-calibrated uncertainty between features or latent representations to enhance interpretability.
Highlights
- 🎉 Two papers, InfoSEM and Geometric Hyena, are accepted to ICML2025!
- 🎉 Our paper on Bayesian active learning with molecular foundation models is accepted to Cheminformatics and GEM@ICLR2025!
- I presented "Bayesian Deep Learning: Priors, Likelihoods, and Representations" at Microsoft Research Cambridge.
- I presented "Bayesian Deep Learning: Models and Experimental Design" at Secondmind.ai research seminar.
- I was selected as an AI fellow in Human-Centered AI by DAAD, Germany.
- I gave a lecture about "Priors in Bayesian Deep Learning" in AI for drug discovery (AIDD) school.
- 🎉 I received the Best Reviewer Award from ICML2021!
Interns and Students
- Yanke Li: Bayesian Optimization for Counterfactual Prediction in Perturbation Design, ETH Zürich
- Muhammad Arslan Masood: Bayesian active learning for drug property prediction, Aalto University
Teaching
- Machine Learning for Neuroscience, Imperial College London, 2024
- Machine Learning: Advanced Probabilistic Methods, Aalto University, 2019 – 2022
Reviewing
- NeurIPS, ICML, ICLR, AISTATS, JMLR, AABI